Biography
I am a Machine Learning Scientist at AI VIVO and a Data Scientist at the Wellcome Sanger Institute. My work is at the intersection of computational biology and drug discovery, where I develop deep generative and foundation models for molecules and cells. I specialize in molecule generation and single-cell perturbation modeling using advanced techniques like VAEs, Diffusions, Transformers, and Flow Matchings. I’m passionate about building AI methods that accelerate target discovery and therapeutic design.
Interests
- Deep Generative Models
- Drug Discovery
- Single-Cell Genomics
- Representation Learning
Education
Applied Computer Science & Artificial Intelligence, 2023
Sapienza University of Rome
BSc in Computer Science, 2020
Amirkabir University of Technology
Work Experience
Machine Learning Scientist
AI VIVO
- Develop deep learning and generative models for drug discovery using transformer and flow matching architectures
- Deploy and scale ML pipelines on GCP using PyTorch Lightning and Docker
- Design multi-modal ML pipelines integrating molecular structure and biological assay data
- Maintain scalable pipelines using PyTorch, Lightning, RDKit, and HuggingFace
- Experience with computational chemistry tools like BioSolveIt, AutoDock Vina, and Boltz-2
Data Scientist
Responsibilities include:
- Research assistant at Dr. Mo Lotfollahi’s lab
- Co–first author of CellDISECT, a deep generative model for disentangled cellular representations and in silico perturbation analysis, developed to study perturbation effects across single-cell populations.
- Collaborated on several ML projects in Single-Cell Genomics and Drug Discovery mainly using PyTorch
- Developed task-specific fine-tuning pipelines on generative and transformer models
- Contributed to ongoing projects like CPA: Compositional Perturbation Autoencoder (GitHub)
Computer Vision Engineer
Responsibilities include:
- Delivered >95% accuracy solutions for tasks with limited data (15 images per class)
- Spearheaded development on 5 diverse projects meeting client requirements
- Led individual projects, enhancing development pipelines
Projects
CellDISECT (Cell DISentangled Experts for Covariate counTerfactuals) is a powerful causal generative model that enhances single-cell analysis by disentangling variations, making counterfactual predictions, and achieving flexible fairness.
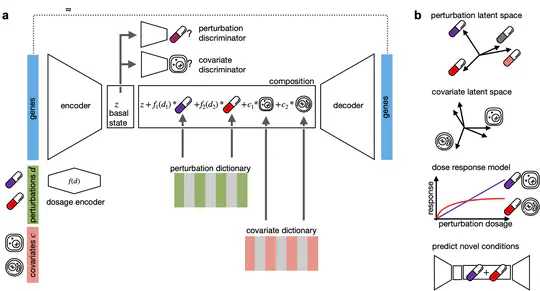
CPA is a deep generative framework to learn effects of perturbations at the single-cell level. It performs OOD predictions of unseen combinations of drugs, learns interpretable embeddings, estimates dose-response curves, and provides uncertainty estimates.
Recent Publications
Quickly discover relevant content by filtering publications.
Recent & Upcoming Talks
Teaching Experience
Teaching Assistant
Sharif University of Technology
- Machine Learning for Bioinformatics (Graduate Course) | Spring 2023
- Prepared teaching material on CNNs & AutoEncoders, designed assignments, and coordinated class contests.
- Introduction to Machine Learning | Fall 2022
- Designed and graded assignments for a class of 150 students, conducted a workshop on Variational AutoEncoders.
Teaching Assistant
Amirkabir University of Technology
- Introduction to Image Processing and Neural Networks | Fall 2022
- Conducted workshops and lectures on OpenCV and Deep Learning for a class of 80 students.
- Advanced Programming with C++ | Spring 2022
- Designed assignments and projects for a class of 90 students, evaluated student submissions.
Accomplishments
- Neural Networks and Deep Learning
- Improving Deep Neural Networks: Hyperparameter Tuning, Regularization and Optimization
- Structuring Machine Learning Projects
- Convolutional Neural Networks
- Sequence Models
Contact
- arianamaani@gmail.com
- Rome, Italy
- DM Me
- Contact Me